Taxon Match Tool
User Manual
- Prepare your species list before matching. For example create columns for each taxon rank (incl. sub, super and infra ranks) and list the species in rows. Remove additional information that is not part of the species name, e.g. erase all notations of sp., cfr., larvae, indet., etc.. The species list can be an excel sheet (example below), plain text or csv file.
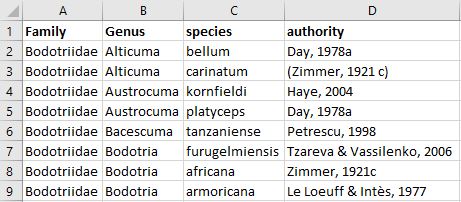
- Click on "choose file" to upload your file. This will open the file explorer on your PC.
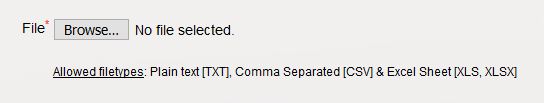
- Select the way you have delimited your taxon names. When you use an MS Excel sheet, this is selected automatically. Use the flag box to indicate whether your first row contains the column names or not.
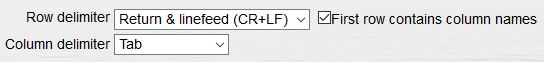
- Use the flag box "Match authority" when you have the authority (=author(s), date) in a separate column and you want to include this as a criterium to find exact matches.


- With the "Match up to" drop-down list you can define to what level in the classification you want to match your list with. For example if your taxon list includes columns for higher ranks such as Class, Order, Family, Genus etc. you can include these in the match query. In this way you can verify your own classification.

If you only have one column with taxon names, so no classification, then you indicate match up to "ScientificName".
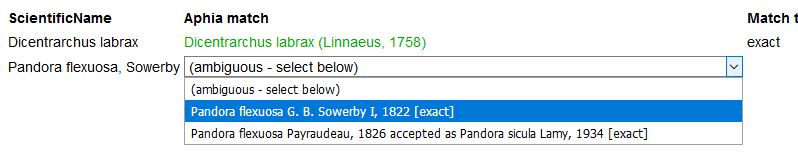
When the taxon name has a homonym, you will be able to choose the right one from a pick list. To avoid ambiguous matches with homonyms in the first place, you can limit your match query to a particular higher taxon, e.g. only within Porifera, Copepoda, etc.. Enter the first characters of the taxon name and use the pick-list.

- Finally, before you click on
 also indicate in the output boxes which data you would like the system to return. AphiaID is the unique identifier of WoRMS; TSN is the unique identifier of ITIS; the valid names option lists the valid names in a separate column (this is important when your list includes unaccepted names or spelling variations). You can also ask to receive the full classification, authority and quality status flags. The output format is in txt, xls or SGML.
also indicate in the output boxes which data you would like the system to return. AphiaID is the unique identifier of WoRMS; TSN is the unique identifier of ITIS; the valid names option lists the valid names in a separate column (this is important when your list includes unaccepted names or spelling variations). You can also ask to receive the full classification, authority and quality status flags. The output format is in txt, xls or SGML.

When you click on
 you will get the following window, were you can fix possible homonyms
you will get the following window, were you can fix possible homonyms

You can choose what the output format of your file will be (for example XLSX) and then click on



- There are several types of matches:
- exact all characters match exactly
- exact_subgenus an exact match, but including the subgenus
- exact_replaced an exact match on a deleted or quarantined name, but returning it's replacement AphiaID & name
- phonetic sounds similar as, despite minor differences in spelling (soundex algorithm)
- near_1 perfect match, except for one character. This is a quite reliable match
- near_2 good match, except for two characters. This needs an extra check
- near_3 good match, except for three characters. This definitely needs an extra check
- match_quarantine match with a name that is currently in quarantine. Any name that has been used in the literature should in principle not be quarantined. So best to contact the WoRMS DMT about this
- match_deleted this is a match with a name that has been deleted and no alternative is available. Please contact the WoRMS DMT when you come across this.